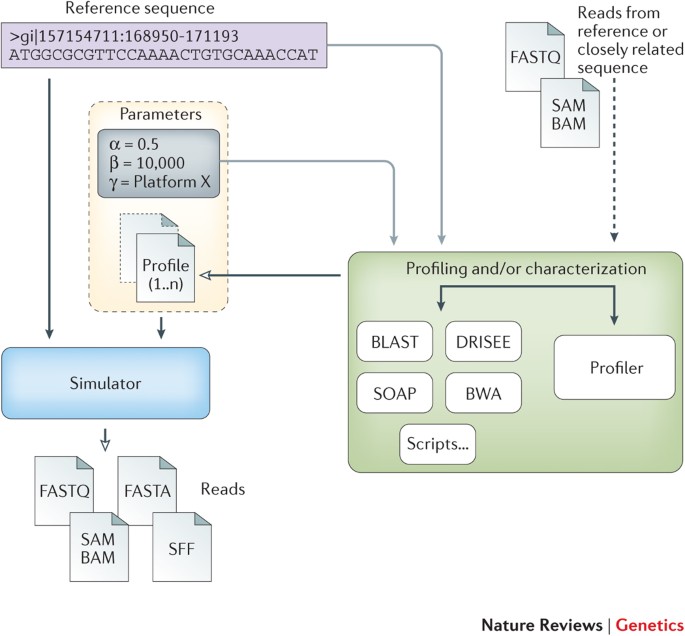
Next-generation sequencing NGS simulators try to imitate the real sequencing process as closely as possible by considering all the steps that could influence the characteristics of the reads. It will facilitate the evaluation and development of new analysis tools for the third-generation sequencing data.

Instead it uses the available sequencing technology nucleotide composition and read length to generate reads from scratch.
Art a next generation sequencing read simulator. ART is a set of simulation tools that generate synthetic next-generation sequencing reads. This functionality is essential for testing and benchmarking tools for next-generation sequencing data analysis including read alignment de novo assembly and genetic variation discovery. ART generates simulated sequencing reads by emulating the.
PDF ART is a set of simulation tools that generate synthetic next-generation sequencing reads. This functionality is essential for testing and. ART can be used to test or benchmark a variety of method or tools for next-generation sequencing data analysis including read alignment de novo assembly SNP and structure variation discovery.
ART was used as a primary tool for the simulation study of the 1000 Genomes Project. ART is a set of simulation tools that generate synthetic next-generation sequencing reads. This functionality is essential for testing and benchmarking tools for next-generation sequencing data analysis including read alignment de novo assembly and genetic variation discovery.
ART generates simulated sequencing reads by emulating the sequencing process with built-in. ART can perform regular genome sequencing simulation as well amplicon sequencing simulation. ART is implemented in C with optimized algorithms and is highly efficient in read simulation.
ART outputs reads in the FASTQ format and alignments in the ALNMAP andor SAM format. ART can also generate alignments in UCSC BED file format. ART is a set of simulation tools that generate synthetic next-generation sequencing reads.
This functionality is essential for testing and benchmarking tools for next-generation sequencing data analysis including read alignment de novo assembly and genetic variation discovery. 3 mate-pair read simulation art_illumina -sam-i referencefa -mp-l 50 -f 20 -m 2500 -s 50 -o matepair_dat 4 amplicon sequencing simulation with 5 end single-end reads art_illumina -amp-sam-na-i amp_referencefa -l 50 -f 10 -o amplicon_5end_dat 5 amplicon sequencing simulation with paired-end reads. GemSIM or General Error-Model based SIMulator is a next-generation sequencing simulator capable of generating single or paired-end reads for any sequencing technology compatible with the generic formats SAM and FASTQ including Illumina and Roche454.
A next-generation sequencing read simulator Bioinformatics 28 4. An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. PaSS is an effective sequence simulator for PacBio sequencing.
It will facilitate the evaluation and development of new analysis tools for the third-generation sequencing data. In the Next Generation Sequencing NGS simulation you will obtain a hair sample from an ancient man from Greenland extract his DNA and perform DNA sequencing. The NGS technology uses second-generation DNA sequencing where users perform sequencing of many samples at the same time also known as massive parallel sequencing.
Next-generation sequencing NGS simulators try to imitate the real sequencing process as closely as possible by considering all the steps that could influence the characteristics of the reads. Mason A Read Simulator for Second Generation Sequencing Data Holtgrewe M. 2010 Mason A Read Simulator for Second Generation Sequencing Data.
Technical Report FU Berlin. PDF - Submitted Version 338kB. We present a read simulator software for Illumina 454 and Sanger reads.
Read simulators can fulfill this requirement but are often criticized for limited resemblance to true data and overall inflexibility. We present NEAT NExt-generation sequencing Analysis Toolkit a set of tools that not only includes an easy-to-use read simulator but also scripts to facilitate variant comparison and tool evaluation. Simulating Sequence Reads from a Reference Genome with wgsim 1 minute read wgsim is a tool within the SAMtools software package that allows the simulation of FASTQ reads from a FASTA reference.
It can simulate diploid genomes with single nucleotide polymorphisms SNP and insertiondeletion indels and create reads with uniform substitution sequencing errors. Next-generation sequencing technologies are fueling a wave of new diagnostic tests. Progress on a key set of nine research challenge areas will help generate the knowledge required to advance effectively these diagnostics to the clinic.
Phylogeny guided Simulator for Tumor Evolution PSiTE PSiTE is a phylogeny guided simulator for tumor evolution. It first simulates somatic variants Single Nucleotide Variants SNVs and Copy Number Variants CNVs along the history of tumor evolution and then generates Next Generation Sequencing NGS data for tumor samples. On the other hand XS read simulator doesnt need any reference sequence to generate reads.
Instead it uses the available sequencing technology nucleotide composition and read length to generate reads from scratch. Next decide whether the reads should be simulated from one or several organisms.