Transcription is regulated through the binding of transcription factor proteins to specific cis -level regulatory sites in the DNA. DNase-seq footprinting enables the prediction of genome-wide binding sites for hundreds of TFs simultaneously.
This track shows probable binding sites of the specified transcription factors TFs in the given cell types as determined by chromatin immunoprecipitation followed by high throughput sequencing ChIP-Seq.
Encode transcription factor binding sites. To predict genome-wide binding sites for hundreds of transcription factors TFs simultaneously. Analyze data from the ENCODE consortium to create a resource of footprints in 27 human tissues demonstrating associations of tissue-specific TF occupancy with gene regulation and disease risk. Funk et al 2020 Cell Reports 32 108029.
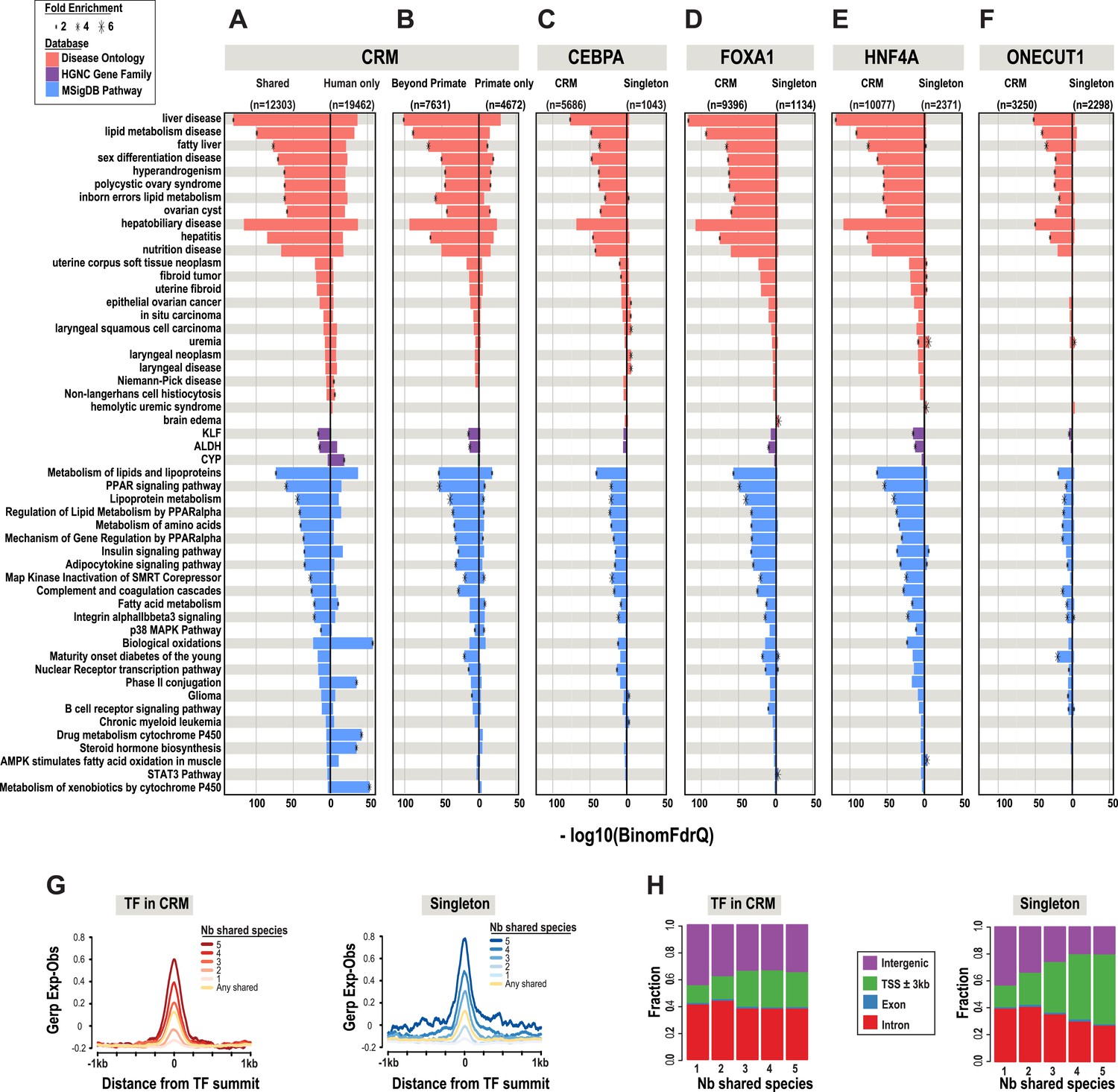
Atlas of Transcription Factor Binding Sites from ENCODE DNase Hypersensitivity Data across 27 Tissue Types Characterizing the tissue-specific binding sites of transcription factors TFs is essential to reconstruct gene regulatory networks and predict functions for non-coding genetic variation. Characterizing the tissue-specific binding sites of transcription factors TFs is essential to reconstruct gene regulatory networks and predict functions for non-coding genetic variation. DNase-seq footprinting enables the prediction of genome-wide binding sites for hundreds of TFs simultaneously.
Despite the public availability of high-quality DNase-seq data from hundreds of samples a. Characterizing the tissue-specific binding sites of transcription factors TFs is essential to reconstruct gene regulatory networks and predict functions for non-coding genetic variation. DNase-seq footprinting enables the prediction of genome-wide binding sites for hundreds of TFs simultaneously.
Despite the public availability of high-quality DNase-seq data from hundreds of samples a comprehensive up-to. Atlas of Transcription Factor Binding Sites from ENCODE DNase Hypersensitivity Data across 27 Tissue Types. 1679 sets of genes with transcription factor binding evidence in transcription factor.
Our workflow then scans footprints for 1530 sequence motifs to predict binding sites for 1515 human transcription factors. We tested our workflow using 21 DNase-seq experiments of lymphoblastoid cell lines generated by the ENCODE project. ChIP-seq combines chromatin immunoprecipitation with DNA sequencing to infer the possible binding sites of DNA-associated proteins.
The ENCODE consortium has developed two analysis pipelines to study two different classes of protein-chromatin interactions. The transcription factor ChIP-seq TF ChIP-seq pipeline described here is suitable for proteins that are expected to bind in a punctate. 21 Zeilen The encoded protein binds the octamer sequence 5-ATTTGCAT-3 a.
Mapping genome-wide binding sites of all transcription factors TFs in all biological contexts is a critical step toward understanding gene regulation. The state-of-the-art technologies for mapping transcription factor binding sites TFBSs couple chromatin immunoprecipitation ChIP with high-throughput sequencing ChIP-seq or tiling array hybridization ChIP-chip. The goal of the ENCODE-DREAM in vivo Transcription Factor Binding Site Prediction Challenge is twofold.
We aim to identify the best performing model for predicting positional in vivo TF binding maps across cell types and tissues. Transcription Factor Binding Sites by ChIP-seq from ENCODEStanfordYaleUSCHarvard Release 2 14 May 2012. RNA Subcellular CAGE Localization from ENCODERIKEN Release 3 02 May 2012.
RNA-seq from ENCODECaltech Release 3 19 Apr 2012. Replication Timing by Repli-chip from ENCODEFSU. If you have questions about the Genome Browser track associated with this data contact ENCODE mailtogenomesoeucscedu.
This track shows probable binding sites of the specified transcription factors TFs in the given cell types as determined by chromatin immunoprecipitation followed by high throughput sequencing ChIP-Seq. Included for each cell type is the input signal. ENCODE Transcription Factor Binding.
Transcription is regulated through the binding of transcription factor proteins to specific cis -level regulatory sites in the DNA. The nature of this regulation depends on the transcription factor. The richness of the ENCODE ChIP-seq data provides an unprecedented opportunity to understand the relationship between transcription factor binding sites and nucleosome positioning.
ENCODE discovers many new transcription-factor-binding-site motifs and explores their properties. These data raise the hypothesis that rare genetic variation at transcription factor binding sites TFBSs might contribute to local DNA methylation patterning. In this work by combining blood genome-wide DNA methylation profiles whole genome sequencing-derived SNVs from 247 unrelated individuals along with 133 predicted TFBS motifs derived from ENCODE ChIP-Seq data we observed an association between the disruption of binding sites.
To enable low-cost high-throughput generation of cistrome and epicistrome maps for any organism we developed DNA affinity purification sequencing DAP-seq a transcription factor TF-binding site TFBS discovery assay that couples affinity-purified TFs with next-generation sequencing of a genomic DNA library. The method is fast inexpensive and more easily scaled than chromatin.
